Section: New Results
Assessing and tuning brain decoders: cross-validation, caveats, and guidelines
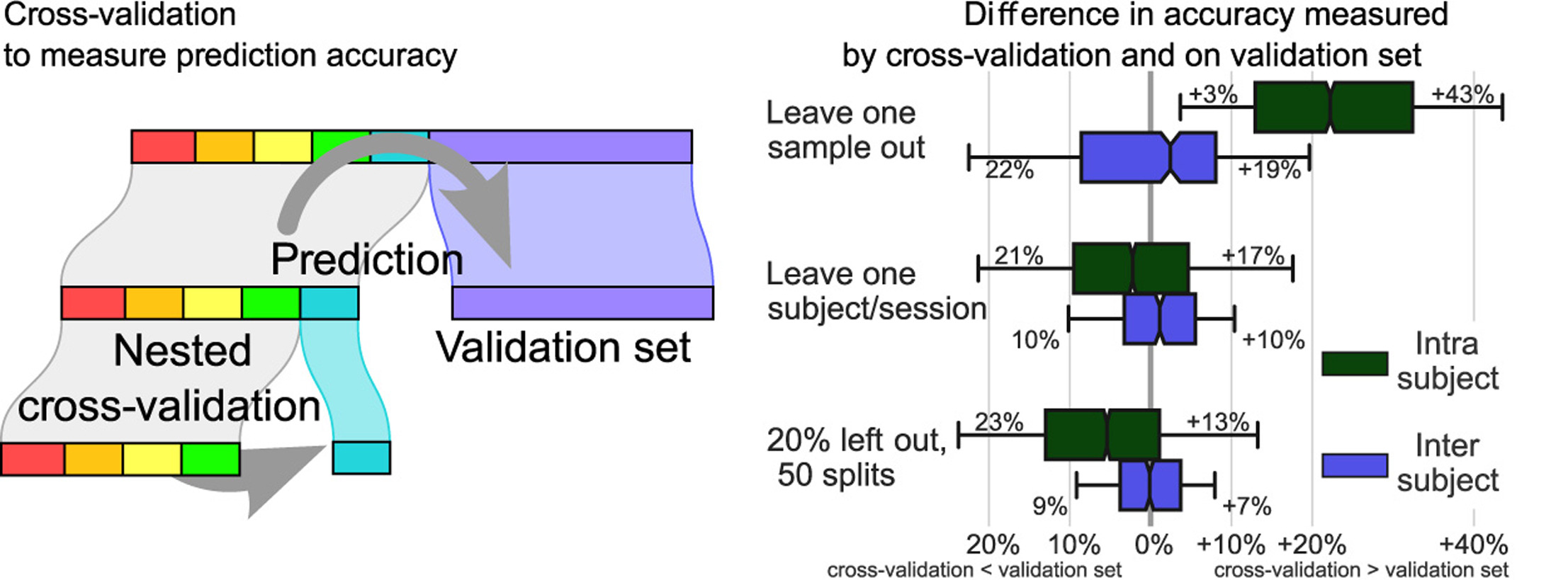
Decoding, ie prediction from brain images or signals, calls for empirical evaluation of its predictive power. Such evaluation is achieved via cross-validation, a method also used to tune decoders' hyper-parameters. This paper is a review on cross-validation procedures for decoding in neuroimaging. It includes a didactic overview of the relevant theoretical considerations. Practical aspects are highlighted with an extensive empirical study of the common decoders in within-and across-subject predictions, on multiple datasets –anatomical and functional MRI and MEG– and simulations. Theory and experiments outline that the popular " leave-one-out " strategy leads to unstable and biased estimates, and a repeated random splits method should be preferred. Experiments outline the large error bars of cross-validation in neuroimaging settings: typical confidence intervals of 10%. Nested cross-validation can tune decoders' parameters while avoiding circularity bias. However we find that it can be more favorable to use sane defaults, in particular for non-sparse decoders.
|
See Fig. 9 for an illustration and [16] for more information.